The genetic code is the information encoded within the mRNA sequence that is translated into proteins by living cells. The codon table is shown. 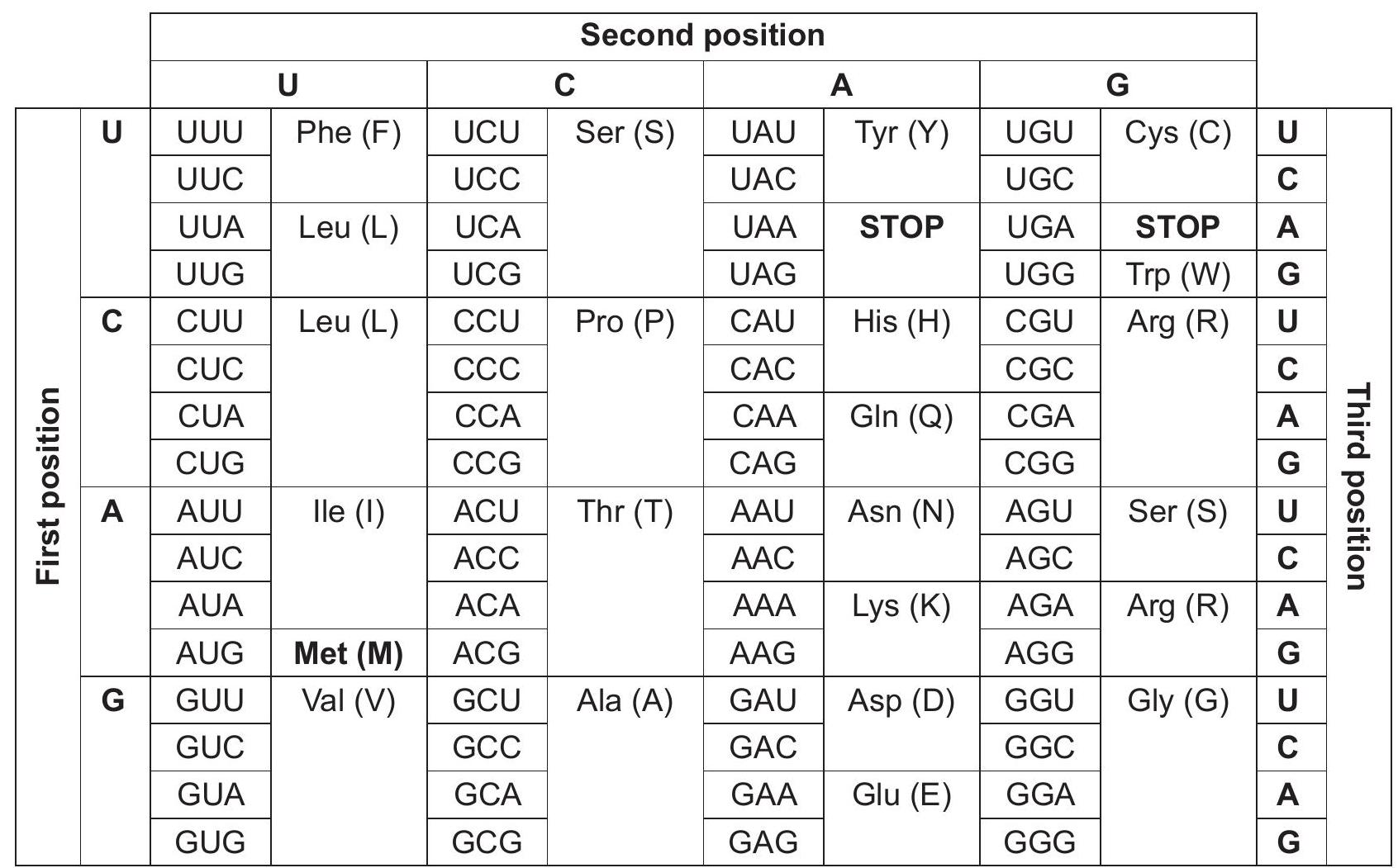 The first part of the cytochrome c protein sequence alignment of mold fungus (Neurospora), horse (Equus), human (Homo), corn (Zea) and rice (Oryza) is shown using the amino acids as a one letter code. Neurospora
Equis
Homo
Zea
Oryza
MGFSAGDSKKGANLFKTRCAQCHTLEEGGGNKIGPALHGLFGRKTGSVDGYAYTDA
--------MGDVEKGKKIFVQKCAQCHTVEKGGKHKTGPNLHGLFGRKTGQAPGFSYTDA
--------MGDVEKGKKIFIMKCSQCHTVEKGGKHKTGPNLHGLFGRKTGQAPGYSYTAA
MASFSEAPPGNPKAGEKIFKTKCAQCHTVDKGAGHKQGPNLNGLFGRQSGTTAGYSYSAG
MASFSEAPPGNPKAGEKIFKTKCAQCHTVDKGAGHKQGPNLNGLFGRQSGTTPGYSYSTA
The first part of the cytochrome c protein sequence alignment of mold fungus (Neurospora), horse (Equus), human (Homo), corn (Zea) and rice (Oryza) is shown using the amino acids as a one letter code. Neurospora
Equis
Homo
Zea
Oryza
MGFSAGDSKKGANLFKTRCAQCHTLEEGGGNKIGPALHGLFGRKTGSVDGYAYTDA
--------MGDVEKGKKIFVQKCAQCHTVEKGGKHKTGPNLHGLFGRKTGQAPGFSYTDA
--------MGDVEKGKKIFIMKCSQCHTVEKGGKHKTGPNLHGLFGRKTGQAPGYSYTAA
MASFSEAPPGNPKAGEKIFKTKCAQCHTVDKGAGHKQGPNLNGLFGRQSGTTAGYSYSAG
MASFSEAPPGNPKAGEKIFKTKCAQCHTVDKGAGHKQGPNLNGLFGRQSGTTPGYSYSTA
State the bioinformatics tool used to obtain the alignment.
BLASTp ✓
State the meaning of the dash (-) in the alignment.
gap/no amino acid present «for cytochrome c in that organism in that position» OR protein is shorter ✓
Identify the longest amino acid sequence where there are no differences amongst the five genera.
GLFGR ✓
Suggest, with a reason, whether the DNA coding for the amino acid sequence identified in (i) must be identical for the five genera.
Describe briefly how the cladogram was obtained.
ALTERNATIVE 1 a. alignment used to quantify differences and similarities ✓ b. algorithms for cladograms OR named algorithms ✓ c. selection of best model ✓ ALTERNATIVE 2 d. comparing amino acid sequences between organisms ✓ e. more similar sequences correspond to closer evolutionary links/OWTTE OR number of differences indicate number of mutations accumulated «over time» OR «if mutation rate is assumed to be constant», more mutations imply further evolutionary distance ✓ f. length of lines/position of nodes established by the number of differences/mutations between organisms ✓

