Practice D2.2 Gene expression (HL) with authentic IB Biology exam questions for both SL and HL students. This question bank mirrors Paper 1A, 1B, 2 structure, covering key topics like cell biology, genetics, and ecology. Get instant solutions, detailed explanations, and build exam confidence with questions in the style of IB examiners.
Define epigenetics and epigenome, and briefly describe their significance in multicellular organisms.
Describe how methylation of the promoter region in DNA impacts gene expression.
Outline one example of environmental effects on gene expression, including the mechanism by which this occurs.
Explain the regulation of transcription by transcription factors that bind to the promoter sequence of DNA.
The function of silencer DNA sequences is to:
Which process involves the removal of introns?
Outline the consequences of genomic imprinting in lion-tiger hybrids, explaining the origin of size differences between ligers and tigons.
Describe how environmental pollutants, such as air pollution, can influence the epigenetic regulation of immune response genes.
Outline the mechanism by which oestrogen regulates gene expression related to endometrial development and maintenance in the uterine cycle.
In the image below, the left panel shows changes in global DNA methylation levels (%) in somatic cells over three generations ( to ) following an environmental stimulus in the generation. The right panel displays the expression level of a phenotypic trait over six generations ( to ), highlighting how the phenotype changes across generations despite no changes in the DNA sequence.
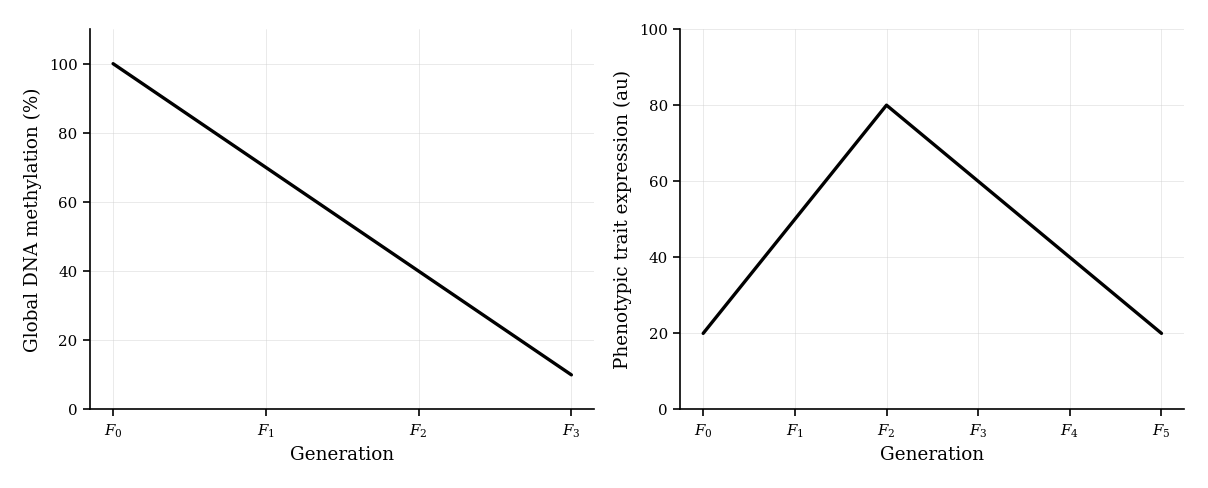
State the trend in DNA methylation observed from to .
Based on the right graph, identify the generation in which the phenotypic trait expression peaks. Suggest a possible epigenetic explanation for this change.
Explain how DNA methylation can be involved in the heritable silencing of gene expression.
Describe how epigenetic changes, such as those shown here, can contribute to phenotypic plasticity without altering the DNA sequence.
Figure showing transcriptional regulation of gene in a eukaryotic cell. DNA bending proteins, transcription factors, mediator proteins, and activators interact with RNA polymerase and control elements to initiate transcription.
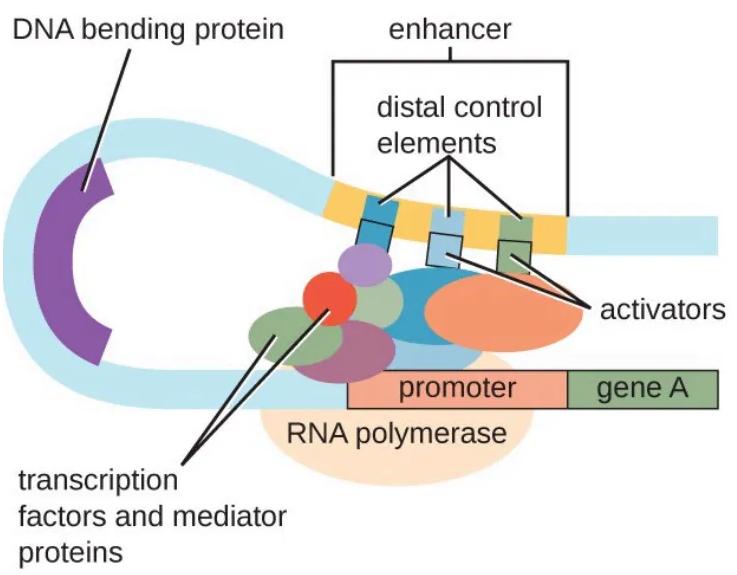
State the function of the promoter shown in the Figure.
Explain the role of transcription factors and mediator proteins in this diagram.
Describe how the enhancer and its associated proteins regulate the transcription of gene .
Suggest one reason why this regulatory mechanism allows differential gene expression in different cell types.
Which of the following sequences acts to increase transcription when bound by an activator?
During transcription, which of the following best describes the role of RNA polymerase?
Promoters are non-coding regions in DNA. What is the role of a promoter?